
My Research
Research Interest
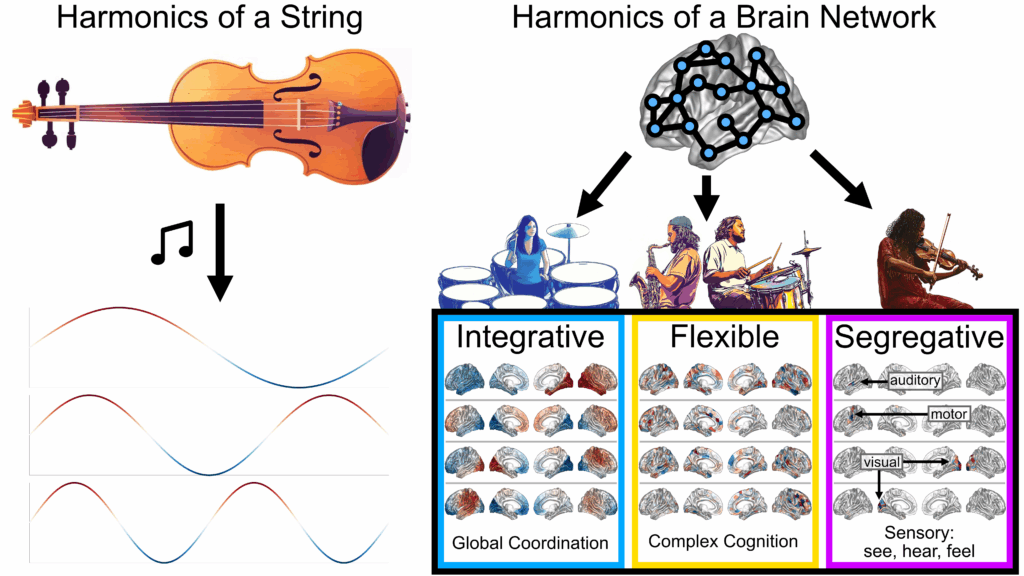
My work applies spectral graph theory to decompose the brain network into
spatial patterns, known as “brain harmonics,” to better understand the brain’s structural and functional networks in development, aging, and disease. I have found that these harmonics form distinct regimes related to integration and segregation in the brain and disentangle subject-specific from shared features.
As an analogy, if the brain were an orchestra, then the structural harmonics describe different orchestral sections that join together to create the music of brain function. Here’s a my performance at a GradSlam competition using this analogy:
Education



PhD in Bioengineering at UCSF and UC Berkeley
Major Area of Focus: Computational Neuroscience
UCSF Master’s of Biomedical Imaging

Major: Animal Biology
Minors: Professional Writing, Sociology
Current Projects
Modeling structural network directionality with the higher-order network diffusion model
Diffusion MRI tractography is widely used to estimate structural connectivity (SC) between brain regions in vivo, but it lacks directional information about white matter pathways. I have used a higher-order network diffusion model to estimate the asymmetry in SC that best explains fMRI brain activity. Using the CoCoMac open-source database as a ground-truth for directionality in non-human primates, I found that my method can accurately recover SC directionality using fMRI in 9 non-human primates.
Filtering out the effects of network diffusion in the brain’s functional signals
It is commonly understood that brain regions perform regionally specific computations that are then transmitted globally through the whole-brain white matter network. However, the success of network diffusion models for predicting the brain’s functional connectivity suggests that a significant portion of recorded brain signal is attributable to passive diffusion through the structural network. We can therefore interpret the global signal as a convolution of an innovation signal from novel regional computations with a passive signal merely flowing through the network. Since the innovation signal contains the information of interest, there is a need to deconvolve the innovation signal from the passive signal in functional brain recordings. Here, we estimate the innovation signal in functional MRI by using the Network Diffusion Model (NDM) to measure the amount of global signal diffusion in data from 64 Human Connectome Project (HCP) subjects, then invert the NDM to perform a Network Diffusion Deconvolution (HONeD deconvolution). We show that the estimated innovation signal has different BOLD power gradients, forms a more segregated functional network, reorganizes resting-state networks, and constrains the signal in a motor task to the expected motor regions. Overall, HONeD deconvolution highlights a promising new approach to study fMRI and connectivity.
Other ongoing projects
- Tracking changes in brain harmonics across lifespan development
- Evaluating the properties of the brain’s complex Laplacian harmonics
- Using HONeD deconvolution to assess the effect of a mindfulness intervention in adolescents (Collaborator: Dr. Tony Yang)
- Studying how brain harmonics change during high dose psychedelics (Collaborators: Robin Carhart-Harris and Lorenzo Pasquini)
Select Publications
Sipes, B. S., Nagarajan, S. S., & Raj, A. (2024). Integrative, segregative, and degenerate harmonics of the structural connectome. Communications biology, 7(1), 986.
Sipes, B. S., & Raj, A. (2025, April). Deconvolving Passive Diffusion on the Structural Network from Functional Brain Signals. In 2025 IEEE 22nd International Symposium on Biomedical Imaging (ISBI) (pp. 1-5). IEEE.
Tymofiyeva, O., Sipes, B. S., Luks, T., Hamlat, E. J., Samson, T. E., Hoffmann, T. J., … & Yang, T. T. (2024). Interoceptive brain network mechanisms of mindfulness-based training in healthy adolescents. Frontiers in Psychology, 15, 1410319.
Raj, A., Sipes, B. S., Verma, P., Mathalon, D. H., Biswal, B., & Nagarajan, S. (2024). Spectral graph model for fMRI: A biophysical, connectivity-based generative model for the analysis of frequency-resolved resting-state fMRI. Imaging Neuroscience, 2, 1-24.
Previous Research
Assistant Specialist. Dr. Olga Tymofiyeva & Dr. Tony Yang, UCSF
Research Volunteer. Dr. Adam Gazzaley & Dr. Courtney Gallen, Neuroscape UCSF
Research Assistant. Dr. Brenda McCowan, UC Davis School of Veterinary Medicine
Assistant Team Leader/Research Assistant. Dr. Karen Bales, UC Davis CNPRC
